Paroxysmal Atrial Fibrillation Events Detection from Dynamic ECG Recordings: The 4th China Physiological Signal Challenge 2021
Invitation Letter (in English) Invitation Letter (in Chinese)
Welcome to join CPSC 2021 Wechat Group for free discussion and information sharing. Please add the conference wechat and remark with "Please invite me into the Challenge Wechat Group"

The final challenge scores of CPSC 2021 are shown as below,
Part I-Scores of the Official Entries
 Please download the codes via the specific Team Name in the score list.
Please download the codes via the specific Team Name in the score list.
| Name | Affiliation | Team Members | Test_I | Test_II | Average |
| usstmed | University of Shanghai for Science and Technology | Wenjie Cai, Fanli Liu, Bolin Xu, Xuan Wang, Yufeng Ji | 2.0629 | 4.3921 | 3.2275 |
| CeZIS | 1.VSL Software, a.s., Košice, Slovakia; 2.Pavol Jozef Šafárik University in Košice, Slovakia | Peter Bugata1, Peter Bugata Jr.1, David Gajdos1, David Hudak1,Vladimira Kmecova1, Monika Stankova1, Lubomir Antoni2, Erik Bruoth2, Simon Horvat2, Richard Stana2, Alexander Szabari2, Gabriela Vozarikova2 | 2.1128 | 3.5716 | 2.8422 |
| UNIWA | University of West Attica, Greece | Lampros Kokkalas, Nicolas A. Tatlas, Stelios M. Potirakis | 2.0144 | 3.3833 | 2.6989 |
| lingshui_BME | 1.Dalian University of Technology; 2.RWTH Aachen University | Yating Hu1, Tengfei Feng2, Hong Tang1 | 1.8754 | 3.5116 | 2.6935 |
| Taozi | Tianjin Medical University, Beihang University | Jingsu Kang, Hao Wen | 1.9972 | 3.0907 | 2.5440 |
| Metformin-121 | National Taiwan University; Academia Sinica; Taiwan Artificial Intelligence Academy Foundation | Tsai-Min Chen, Yi-Dar Tang, Huan-Hsin Tseng, Wei Luok Ngu, Le-Yin Hsu, Miao-Chen Chiang, Yu-Te Ku, Ming-Yi Hong, Yu Tsao | 1.6277 | 2.6649 | 2.1463 |
| MVTECH | Shanghai Medical Vision Technology Co. Ltd. | Yang Hou, Jinlei Li | 1.7966 | 2.4346 | 2.1156 |
| FUDU_Car | Fudan University | Sen Liu, Ya‘nan Wang, Haijun Jia | 1.9147 | 1.9473 | 1.9310 |
| WHS | 1.Central South University 2.China University of Geosciences | Lebing Pan, Jiechen Tang | 1.8585 | 1.9236 | 1.8911 |
| Muhammad Uzair Zahid | 1. Tampere University, Finland; 2. Qatar University, Qatar | Muhammad Uzair Zahid1, Mustafa Serkan Kiranyaz2, Moncef Gabbouj1 | 1.8192 | 1.9469 | 1.8831 |
| DaBin | Beijing University of Technology | Fengya Liu, Rui Yu, Shuicai Wu, Guangyu Bin,Zhuhuang Zhou ,Qian Wang | 1.8352 | 0.9728 | 1.4040 |
| Lastone | King's College London, UK | Xinqi Bao, Fenghe Hu | 0.9616 | 1.6430 | 1.3023 |
| BSU工程小分队 | Beijing Sport University | Kuan Tao, Lixin Sun | 1.2384 | 1.1710 | 1.2047 |
| CUTCM | City University of Hong Kong | Marshall | 0.6706 | 1.3854 | 1.0280 |
| AZ-unet | AstraZeneca | Hannes Whittingham, Long Luu | 1.1168 | 0.7788 | 0.9478 |
| AIBI_LAB | Ludong University | Shuhong Wei, Yipeng Wang, Yu Ji, Yinhao Sun | 0.7837 | 1.1006 | 0.9422 |
| CPSC_eie | Zhejiang University of Technology, China | Xinyuan Ying, Qing Pan, Ziyou Zhang | 0.8555 | 0.9629 | 0.9092 |
| Baseline | / | / | 0.6819 | 0.6485 | 0.6652 |
Part II-Scores of the Unofficial Entries
| Name | Affiliation | Team Members | Test_I | Test_II | Average |
| Shinall Technology | Shinall Technology | Min Chen,Kui Dong | 1.9940 | 3.8858 | 2.9399 |
| busywork | Shanghai National Group Health Technology Co., Ltd | Jie Y. | 1.5806 | 3.1973 | 2.3890 |
| ECG-Go | Beijing University of Technology | Minggang Shao, Zhuhuang Zhou, Shuicai Wu | 1.6729 | 2.3462 | 2.0096 |
| Proton | Hangzhou Proton Technology Co.,Ltd. | Yamin Liu, Hanshuang Xie, Qineng Cao, Jiayi Yan, Fan Wu, Huaiyu Zhu, Yun Pan | 1.6491 | 2.0718 | 1.8605 |
| TeleCardio | SHANGHAI YUANXIN MEDICAL TECHNOLOGY CO., LTD; Xinghua City People’s Hospital | Tammy Liu, Hao Sun, Qingchun Zhang, Aiguo Wang | 1.7019 | 2.0097 | 1.8558 |
| Zoncare | Wuhan Zoncare Bio-medical Electronics Co., Ltd. | Jiabing Zhu, Quan Fang, Hongling Zhu | 1.3849 | 1.8276 | 1.6063 |
| pancake | Sichuan University | Haoyue Lu, Yiping Huang | 0.8996 | 2.0687 | 1.4842 |
| ECG_SMU | Southern Medical University | Jiewei Lai, Huixin Tan, Zuo Wang, Yue Zhang | 1.8469 | 1.0172 | 1.4321 |
| Cardiac Monitoring | Ludong University | Junwei Fan, Xiangyi Zhu, Zhenxing Zhang | 1.6401 | 1.0103 | 1.3252 |
| NEU-ECG | College of Information Science and Engineering, Northeastern University | Youhe Huang, Songbai Liu, Yi Yang, Ziyang Ren, Rui Tao, Shuang Wen | 1.3019 | 1.1831 | 1.2425 |
| Yunpu St. | CVTE Inc. | Yunju Ma, Wei Zhao, Jing Hu, Zhenqi Li, Fei Wang | 0.7504 | 0.8162 | 0.7833 |
| CSU_ECG | Central South University | Yuxia Guan | 0.3899 | 0.8353 | 0.6126 |
| Baseline | / | / | 0.6819 | 0.6485 | 0.6652 |
Notes
 [2021-5-17] Announcement of the training set updating
[2021-5-17] Announcement of the training set updating
To avoid ambiguity in annotation, the standard for annotation of AF events in the challenge is set as follows: the ECG episodes labeled as 'AFIB' or 'AFL' must contain no less than 5 heart beats; the non-AF episodes must also contain no less than 5 heart beats.
According to the feedback from the contestants, there are still cases where the heart beats of the AF episode or the pause between adjacent AF episodes are less than 5 in the annotation of the training set. In order to make the challenge more rigorous, we revised the data and annotations in the above situation. The id numbers of the revised records are summarized in the attached REVISED_RECORDS file.
Wish the contestants all the best and achieve good results. We also welcome the feedback of the data and annotation errors and questions from the teams.
Please download the updated training set from:
training_I
training_II
[2021-5-8]: The new version of the training set has been released after re-checking, rectifying some avoidable annotation errors due to bugs in data generation. Training_I has been expanded from 716 to 730 ECG recordings. Please download and use the new training set as soon as possible. All download links on the challenge webpage has been updated simultaneously.
[2021-4-1]: The training set in phase II has already been updated.
The Score List has been indicated as below, more will be updated soon…
| Name | Affiliation | Team Members | Highest Score | Latest Score (updated on October 26) |
| CeZIS | 1.VSL Software, a.s., Košice, Slovakia; 2.Pavol Jozef Šafárik University in Košice, Slovakia | Peter Bugata1, Peter Bugata Jr.1, David Gajdos1, David Hudak1,Vladimira Kmecova1, Monika Stankova1, Lubomir Antoni2, Erik Bruoth2, Simon Horvat2, Richard Stana2, Alexander Szabari2, Gabriela Vozarikova2 | 2.1189 | 2.1112 |
| usstmed | University of Shanghai for Science and Technology | Wenjie Cai, Fanli Liu, Bolin Xu, Xuan Wang, Yufeng Ji | 2.0629 | 2.0629 |
| UNIWA | University of West Attica, Greece | Lampros Kokkalas, Nicolas A. Tatlas, Stelios M. Potirakis | 2.0144 | 1.9808 |
| Shinall Technology | Shinall Technology | Min Chen,Kui Dong | 1.9960 | 1.9412 |
| Taozi | Tianjin Medical University, Beihang University | Jingsu Kang, Hao Wen | 1.9781 | 1.9781 |
| Metformin-121 | National Taiwan University; Academia Sinica; Taiwan Artificial Intelligence Academy Foundation | Tsai-Min Chen, Yi-Dar Tang, Huan-Hsin Tseng, Wei Luok Ngu, Le-Yin Hsu, Miao-Chen Chiang, Yu-Te Ku, Ming-Yi Hong, Yu Tsao | 1.9484 | 1.9484 |
| Cardiac Monitoring | Ludong University | Junwei Fan, Xiangyi Zhu, Zhenxing Zhang | 1.9452 | 1.6401 |
| FUDU_Car | Fudan University | Sen Liu, Ya‘nan Wang, Haijun Jia | 1.9147 | 1.9147 |
| lingshui_BME | 1.Dalian University of Technology; 2.RWTH Aachen University | Yating Hu1, Tengfei Feng2, Hong Tang1 | 1.8785 | 1.8785 |
| WHS | 1.Central South University 2.China University of Geosciences | Lebing Pan, Jiechen Tang | 1.8585 | 1.8585 |
| ECG_SMU | Southern Medical University | Jiewei Lai, Huixin Tan, Zuo Wang, Yue Zhang | 1.8460 | 1.8460 |
| DaBin | Beijing University of Technology | Fengya Liu, Rui Yu, Shuicai Wu, Guangyu Bin,Zhuhuang Zhou ,Qian Wang | 1.8352 | 1.8352 |
| Muhammad Uzair Zahid | 1. Tampere University, Finland; 2. Qatar University, Qatar | Muhammad Uzair Zahid1, Mustafa Serkan Kiranyaz2, Moncef Gabbouj1 | 1.8192 | 1.8192 |
| busywork | Shanghai National Group Health Technology Co., Ltd | Jie Y. | 1.8036 | 1.5806 |
| MVTECH | Shanghai Medical Vision Technology Co. Ltd. | Yang Hou, Jinlei Li | 1.7966 | 1.7966 |
| TeleCardio | SHANGHAI YUANXIN MEDICAL TECHNOLOGY CO., LTD; Xinghua City People’s Hospital | Tammy Liu, Hao Sun, Qingchun Zhang, Aiguo Wang | 1.7019 | 1.7019 |
| ECG-Go | Beijing University of Technology | Minggang Shao, Zhuhuang Zhou, Shuicai Wu | 1.6729 | 1.6729 |
| Lastone | King's College London, UK | Xinqi Bao, Fenghe Hu | 1.6585 | 1.6430 |
| Proton | Hangzhou Proton Technology Co.,Ltd. | Yamin Liu, Hanshuang Xie, Qineng Cao, Jiayi Yan, Fan Wu, Huaiyu Zhu, Yun Pan | 1.6491 | 1.6491 |
| AIBI_LAB | Ludong University | Shuhong Wei, Yipeng Wang, Yu Ji, Yinhao Sun | 1.4974 | 1.3454 |
| NEU-ECG | College of Information Science and Engineering, Northeastern University | Youhe Huang, Songbai Liu, Yi Yang, Ziyang Ren, Rui Tao, Shuang Wen | 1.3646 | 1.3646 |
| Zoncare | Wuhan Zoncare Bio-medical Electronics Co., Ltd. | Jiabing Zhu, Quan Fang, Hongling Zhu | 1.3169 | 1.3169 |
| BSU工程小分队 | Beijing Sport University | Kuan Tao, Lixin Sun | 1.2384 | 1.2384 |
| CUTCM | City University of Hong Kong | Marshall | 1.0861 | 0.8805 |
| pancake | Sichuan University | Haoyue Lu, Yiping Huang | 0.8996 | 0.8996 |
| Yunpu St. | CVTE Inc. | Yunju Ma, Wei Zhao, Jing Hu, Zhenqi Li, Fei Wang | 0.7504 | 0.7504 |
| GZHU-ECG | 1.Guangzhou University; 2.Nanjing University of information and technology | Dandan Peng1, Le Sun2,Fan Wang2 | 0.6214 | 0.6214 |
| CSU_ECG | Central South University | Yuxia Guan | 0.3899 | 0.3899 |
| WEIPA | 1.Soochow University 2.Suzhou Institute of Biomedical Engineering and Technology,Chinese Academy of Sciences | Lirong Wang, Lishen Qiu, Wenqiang Cai, Guo Yunbo, Wenliang Zhu, Miao Zhang, Duoduo Wang, Huimin Zhang, Hui Tang, Rui Bao | 0.1821 | 0.1821 |
| Baseline | / | / | 0.6819 | |
Data Download
Introduction
The 4th China Physiological Signal Challenge 2021 (CPSC 2021) aims to encourage the development of algorithms for searching the paroxysmal atrial fibrillation (PAF) events from dynamic ECG recordings.ECG signal provides an important role in non-invasively monitoring and clinical diagnosis for cardiovascular disease (CVD). AF is the most frequent arrhythmia, but PAF often remains unrecognized[1, 2]. Early screening and early detection of paroxysmal AF are particularly important. It is of great value for AF surgery options, drug intervention, and the diagnosis and treatment of various clinical complications [3].
Although accurate detection of paroxysmal AF is very important, there is currently no algorithm that can efficiently measure the onsets and offsets of AF events in dynamic or wearable ECGs [4]. Previous AF detection algorithms usually focus on the classification of AF rhythm, such as entropy feature-based [5, 6] or machine learning-based methods [7, 8], without the location of onsets and offsets of AF events. Thus, the clinical significance for the personalized treatment and management of AF patients is limited. In clinical applications, other abnormal rhythms can significantly influence the accurate identification of AF rhythm. In this year’s challenge, we focus on the detection of paroxysmal AF events from dynamic ECGs. A new dynamic ECG database containing episodes with totally or partly AF rhythm, or non-AF rhythm was constructed, to encourage the development of the more efficient and robust algorithms for paroxysmal AF event detection.
Challenge Data
Data are recorded from 12-lead Holter or 3-lead wearable ECG monitoring devices. Challenge data provides variable-length ECG records fragments extracted from lead I and lead II of the long-term dynamic ECGs, each sampled at 200 Hz. In order to avoid ambiguity in annotation, an AF event is limited to contain no less than 5 heart beats.The training set in the 1st stage consists of 730 records, extracted from the Holter records from 10 AF patients (5 PAF patients) and 39 non-AF patients (usually including other abnormal and normal rhythms).
The training set in the 2nd stage consists of 706 records from 37 AF patients (18 PAF patients) and 14 non-AF patients.
The test set comprises data from the same source as the training set as well as different data source. We ensure that at least one test subset was collected by a different ECG monitoring system compared with the training set. Same as in previous years, we are not planning to release the test set at any point.
All data is provided in WFDB format and the annotations are standardized according to PhysioBank Annotations (link: https://archive.physionet.org/physiobank/annotations.shtml). The annotation includes the beat annotations (R peak location and beat type), the rhythm annotations (rhythm change flag and rhythm type) and the diagnosis of the global rhythm. Please refer to the example code entry (link: https://github.com/CPSC-Committee/cpsc2021-python-entry ) of the challenge for specific data and label load functions. Note that the flag of atrial fibrillation and atrial flutter (‘AFIB’ and ‘AFL’) in annotated information are seemed as the same type when scoring the method.
Please download the training data from here ( Training Set I and Training Set II).
Scoring
For this year’s Challenge, we developed a new scoring metric that awards the correct detection of paroxysmal AF events. The scoring metric includes two steps:- the first step is to classify the rhythm types: non-AF rhythm (N), persistent AF rhythm (AFf) and paroxysmal AF rhythm (AFp).
- the second step is to locate the onset and end for any AF episode prediction.
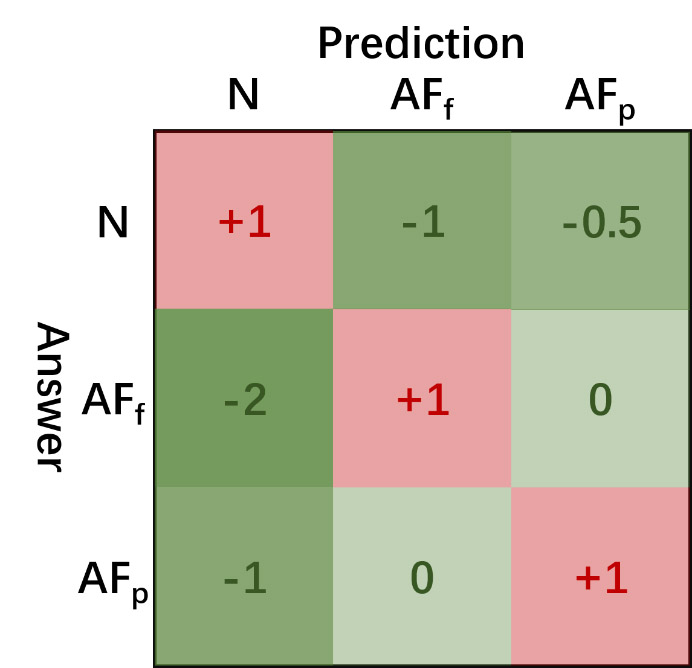
Figure 1. The scoring matrix for the global rhythm classification.
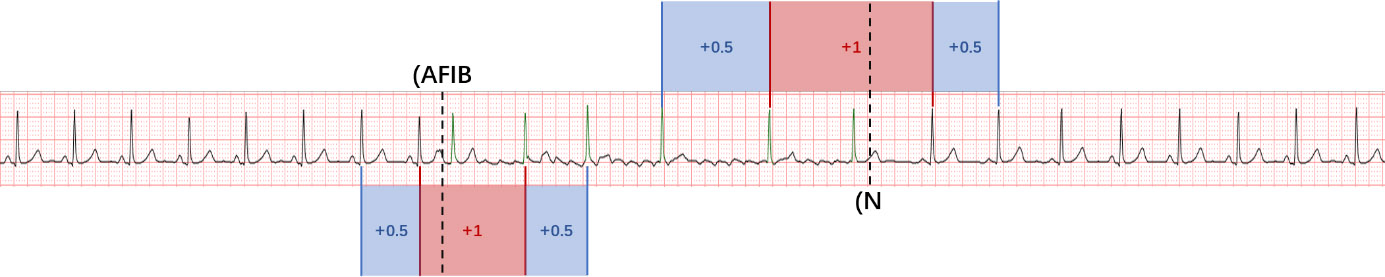
Figure 2. Grading instance for onset and end detections of AF episodes.
Then the detection of onsets and ends of AF episode will be scored (only reward), and the reward is based on the consistence between the detected onsets (or ends) and the annotated answers. Figure 2 gives the detailed explanation. Let Ue be the scores for evaluating the detection of onsets and ends. We reward score Ue with +1 if the detected onset (or end) is within ±1 beat of the annotated position, and score Ue with +0.5 if within ±2 beats. Note that a paroxysmal AF record may contain multiple AF episodes and the onset and end points will be separately scored (rewarded). A weight
 is assigned to Ue, where Ma and Mr are the amount of AF episodes from annotated answers and the predictions, for each record respectively. Over-estimation of AF episodes may cause lower scores.
is assigned to Ue, where Ma and Mr are the amount of AF episodes from annotated answers and the predictions, for each record respectively. Over-estimation of AF episodes may cause lower scores.
The final score U is the sum of Ur and Ue, and is defined as:
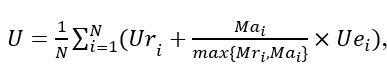
where N is the amount of the test records. The score is calculated for each record and then averaged for the entire test set.
For example, for an ECG record with only one paroxysmal AF episode, if the algorithm classified the record correctly, the first score Ur=1. Then the predicted onset and end of AF episode will be checked. If the both onset and end are within ±1 beat of the annotated position, the second score Ue=2. Thus, the final score U=3.
Sample Submission
A simple example algorithm is provided and may be used as a template for your own submission. Python, MATLAB implementations are available. Similar to last year’s Challenge, teams must submit both the code for their models and the code for training their models (if you are using supervised learning methods).Note that the format of the answer file must follow the entry code. Please save the answer file for each record and name one after the record name. If your development environment is Python, please save the result as a ‘.json’ file. The format is as {‘predict_endpoints’: [[s0, e0], [s1, e1], …, [sm-1, em-2]]. If your development environment is MATLAB, please save the variable, ‘predict_endpoints’, as a ‘.mat’ file.
Preparation and submission instructions
1. Create a private GitHub or Gitlab repository for your code. We recommend cloning our example code and replacing it with your code. Add cpsc-helper as a collaborator to your repository.2. Add your algorithm code to your repository. Like the example code, your code must be in the root directory of the master branch.
3. Do not include extra files that are not required to create and run your code, such as the training data.
4. Follow the instructions for the programming language of your submission.
5. Register through http://www.cpscsub.com/Users/Register. We will clone your repository using the HTTPS URL that ends in .git. On GitHub, you can get this URL by clicking on “Clone or download” and copying and pasting the URL, e.g., https://github.com/CPSC-Committee/cpsc2021-python-entry.git. Please see here (link: https://docs.github.com/en/github/using-git/which-remote-url-should-i-use) for an example.
6. We will put the scores for successful entries on the leaderboard. The leaderboard will publicly show your team name, run time, and score. Note that the limitation of averaged code-run-time should not exceed 1 minute per testing record. If the code running time exceeds the limit, a timeout error will be feedback.
Python-specific instructions
1. Using our Sample Python entry ( https://github.com/CPSC-Committee/cpsc2021-python-entry) as a template, format your code in the following way. Consider downloading this repository, replacing our code with your code, and adding the updated files to your repository.2. requirements.txt: Add Python packages to be installed with pip. Specify the versions of these packages that you are using on your machine. Remove unnecessary packages, such as Matplotlib, that your submitted code does not need.
3. AUTHORS.txt, LICENSE.txt, README.md: Update as appropriate. Please include your authors.
4. entry_2021.py: Update this script to load and run your trained model. While testing, we will firstly run (python entry_2021.py TEST_SET_PATH RESULT_SAVE_PATH) to generate the prediction results for next-step scoring.
5. utils_2021.py: Do not change this script. It is a script containing the functions for the baseline method.
6. score_2021.py: Do not change this script. It is a sample script for reference and help participants test their code in local environment. Another standardized score will be used in the back-end server.
MATLAB-specific instructions
1. Confirm that your MATLAB code compiles and runs in MATLAB R2020B or R2021A.2. Using our sample MATLAB entry ( https://github.com/CPSC-Committee/cpsc2021-matlab-entry) as a template, format your code in the following way. Consider downloading this repository, replacing our code with your code, and adding the updated files to your repository.
3. AUTHORS.txt, LICENSE.txt, README.md: Update as appropriate. Please include your authors. Unfortunately, our submission system will be unable to read your README file to change how we run your code.
4. comp_cosEn.m: Do not edit this script. It extracts extract features from the ECG recordings.
5. qrs_detect.m: Do not edit this script. It extracts the position of the R peak.
6. challenge.m: Update this script to load and run your model weights and any parameters from files in your submission. It takes the input sample path, and returns predict endpoints.
7. Result.m: Update this script to load and run your model weights and any parameters from files in your submission. It takes the input sample name, sample path, save path, and returns predict endpoints and saves to a specific path with a file name as the result of submission.
8. Add your code to the root/base directory of the master branch of your repository.
9. We will download your code, compile it using the MATLAB compiler: Outputting each result (mcc -m challenge.m –a .) and saving to a specific path with a file name as the result of submission (mcc -m Result.m -a .), and run them on backend server.
Awards and Rules
The winner will be selected on the basis of the obtained final U on the hidden test data. The first three for each Event challenging will receive certificates and generous bonuses:• First prize: Certificate plus bonus of RMB 15,000
• Second prize: Certificate plus bonus of RMB 10,000
• Third prize: Certificate plus bonus of RMB 5,000
We welcome all the individual or research group around the world to attend the challenge. To be eligible for the award, please do the following:
(1) Register and submit open-source entries that can be scored before October 25.
(2) Attend ICBEB 2021 (November 15-18, 2021) in Suzhou, China and present your work there.
Please do not submit analysis of this year’s Challenge data to other Conferences or Journals until after ICBEB 2021 has taken place, so the competitors are able to discuss the results in a single forum.
Important Dates
20 March, 2021 -- Challenge openOctober 25, 2021 -- Deadline for team registration to join the challenge
October 25, 2021 -- Deadline for submitting the open-souce challenge entries
November 1, 2021 -- Confirm for submitting the best entry for final scoring
November 15-18, 2021 – Announcement of the winners of CPSC 2021 in ICBEB 2021
Any questions or problems about the Challenge, please feel free to contact chinachallenge_cpsc@icbeb.org.
Challenge Chair:
Chengyu LiuChallenge Committee:
Xingyao WangCaiyun Ma
Xiangyu Zhang
Hongxiang Gao
Songsheng Zhu
International Advisory Chair:
Gari D. CliffordInternational Advisory Co-chairs:
Jianqing LiAiguo Song
Minglong Chen
Kang-Ping Lin
Yuqiu Zhong
International Advisory Committee:
Eddie Ng Yin KweeYi Peng
Zhengtao Cao
Xianzheng Sha
Xingming Guo
Shoushui Wer
Chang Cui
Alistair Johnson
Hosted by:
School of Instrument Science and Engineering, Southeast University, ChinaThe State Key Laboratory of Bioelectronics, Southeast University, China
School of Biomedical Engineering and Information, Nanjing Medical University, China
Supported by:
ICBEB Organizing CommitteeAwards sponsored by:
Nalong Technology Co., Ltd.Reference
[1] G. F. Michaud and W. G. Stevenson, "Atrial Fibrillation," New England Journal of Medicine, vol. 384, no. 4, pp. 353-361, 2021.[2] C. Ma, S. Wei, T. Chen, J. Zhong, Z. Liu, and C. Liu, "Integration of results from convolutional neural network in a support vector machine for the detection of atrial fibrillation," IEEE Transactions on Instrumentation and Measurement, 2020.
[3] R. R. De With et al., "Temporal patterns and short-term progression of paroxysmal atrial fibrillation: data from RACE V," EP Europace, vol. 22, no. 8, pp. 1162-1172, 2020.
[4] H. Baumgartner et al., "2020 ESC Guidelines for the management of adult congenital heart disease: The Task Force for the management of adult congenital heart disease of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC), International Society for Adult Congenital Heart Disease (ISACHD)," European heart journal, vol. 42, no. 6, pp. 563-645, 2021.
[5] C. Liu et al., "A comparison of entropy approaches for AF discrimination," Physiological measurement, vol. 39, no. 7, p. 074002, 2018.
[6] L. Zhao, C. Liu, S. Wei, Q. Shen, F. Zhou, and J. Li, "A new entropy-based atrial fibrillation detection method for scanning wearable ecg recordings," Entropy, vol. 20, no. 12, p. 904, 2018.
[7] V. Kalidas and L. S. Tamil, "Detection of atrial fibrillation using discrete-state Markov models and Random Forests," Computers in biology and medicine, vol. 113, p. 103386, 2019. [8] X. Zhang, J. Li, Z. Cai, L. Zhang, Z. Chen, and C. Liu, "Over-fitting suppression training strategies for deep learning-based atrial fibrillation detection," Medical & Biological Engineering & Computing, vol. 59, no. 1, pp. 165-173, 2021.
